How to Begin Implementing a Molecular Communication in NS3
To stimulate a Molecular Communication project using NS3 it includes the replicate a communication for the nanoscale, in which data is transmitted using molecules instead of electromagnetic waves. Below is a guide to get started:
Steps to Begin Implementing a Molecular Communication Projects Using NS3
Step 1: Understand Molecular Communication
- Key Concepts:
- Transmitter: Encrypts the data in molecules and releases them.
- Medium: The environments by that molecule are broadcast for instance water, air).
- Receiver: It finding and decrypt the molecular signals.
- Noise Sources: It contains the molecular degradation, random broadcast, and external interference.
- Types of Molecular Communication:
- Diffusion-based: Molecules are broadcast through Brownian motion.
- Flow-assisted: Molecules are carried through a medium flow.
- Active Transport: Molecules are transported through biological entities such as molecular motors.
- Performance Metrics:
- Signal-to-noise ratio (SNR).
- Throughput and latency.
- Bit error rate (BER).
Step 2: Set Up NS3 Environment
- Install NS3:
- Download the tool NS3 from the official website.
- Install vital dependencies for sample gcc, Python.
- Add Molecular Communication Extensions:
- NS3 tool doesn’t have natively helps for molecular communication. We may:
- Use the third-party extensions such as Nano-Sim.
- Execute the custom modules for molecular communication.
- NS3 tool doesn’t have natively helps for molecular communication. We may:
- Verify Installation:
- Validate the environment using sample scripts provided in NS3 or Nano-Sim.
Step 3: Design the Molecular Communication System
- Network Components:
- Nanotransmitters: Exude the information for molecules.
- Nanoreceivers: Discover the molecules and decrypt information.
- Propagation Medium: Describe the diffusion and degradation properties.
- Communication Model:
- Select the kind of communication for instance diffusion-based, flow-assisted.
- Molecule Properties:
- Describe the kinds of molecule, size, and chemical elements.
- Data Encoding:
- Select the encrypt system for instance binary concentration encoding.
- Simulation Area:
- Express the 3D replication space and boundary environments.
Step 4: Implement Molecular Communication in NS3
- Create Nodes:
- It can use the node like NodeContainer for built a nanotransmitters and nanoreceivers.
- Implement Propagation:
- Describe the molecule broadcast using diffusion equations or custom broadcast design.
- Develop Communication Protocols:
- Encrypt/decrypt for the information molecules.
- Molecule release and detection for during the synchronization
- Energy Model (Optional):
- It replicates the energy usage for molecule emission and detection.
Step 5: Simulate and Trace
- Set Simulation Parameters:
- Describe the molecule release intervals, broadcast time, and duration for the replication.
- Enable Tracing:
- Store the molecule broadcast paths, receiver finding action, and errors.
- Run the Simulation:
- Use the process for Simulator::Run() for implement the replication.
Step 6: Analyze Results
- Collect Data:
- Calculate the data gathering for SNR, BER, and molecule finding rates.
- Visualize Results:
- Use tools like Python (Matplotlib) or MATLAB for envision.
- Optimize Parameters:
- Research by kinds of molecule for transmitter-receiver distances, and diffusion properties.
Step 7: Document and Present
- Prepare Documentation:
- Save the document has includes the setting, simulation steps, and outcomes.
- Create Visualizations:
- Use the tool like graphs and charts for demonstrate the detection.
Example N3 Implementation Framework
Since NS3 doesn’t have natively helps for molecular communication, here’s a high-level approach for encompassing the NS3:
- Define Molecular Communication Models
- Enhance the classes for:
- Molecule broadcast.
- It behaviors the Nanotransmitter and nanoreceiver.
- Propagation Logic
- Execute the molecule for diffusion using the Brownian motion equations: x(t)=x(0)+2⋅D⋅t⋅N(0,1)x(t) = x(0) + \sqrt{2 \cdot D \cdot t} \cdot N(0,1)x(t)=x(0)+2⋅D⋅t⋅N(0,1) Where:
- DDD: Diffusion coefficient.
- N(0,1)N(0,1)N(0,1): Gaussian random variable.
- Molecule Detection
- Describe the thresholds for molecule findings at the receiver.
- Use Nano-Sim Extension (Optional)
- Incorporate the Nano-Sim, that helps for molecular communication:
Example: Diffusion-Based Molecular Communication
High-Level Pseudo-Code
// Create nanotransmitter and nanoreceiver nodes
NodeContainer nodes;
nodes.Create(2); // Transmitter and Receiver
// Set transmitter behavior
Ptr<Nanotransmitter> transmitter = CreateObject<Nanotransmitter>();
transmitter->SetPosition(Vector(0, 0, 0));
transmitter->SetEmissionRate(100); // Molecules per second
// Set receiver behavior
Ptr<Nanoreceiver> receiver = CreateObject<Nanoreceiver>();
receiver->SetPosition(Vector(10, 0, 0));
receiver->SetDetectionThreshold(5); // Molecules required for detection
// Simulate diffusion
Ptr<MolecularPropagation> propagation = CreateObject<MolecularPropagation>();
propagation->SetMediumProperties(diffusionCoefficient, degradationRate);
propagation->ConnectNodes(transmitter, receiver);
// Run simulation
Simulator::Stop(Seconds(10.0));
Simulator::Run();
Simulator::Destroy();
Molecular Communication Project Ideas
- Diffusion Efficiency:
- It associate the various broadcast models for instance pure diffusion vs. flow-assisted.
- Error Reduction:
- Execute the advanced encode/decode schemes for decrease the BER.
- Energy Efficiency:
- Examine the energy usage for nanotransmitters.
- Receiver Sensitivity:
- Investigate the effect of various finding for thresholds.
- Multi-Node Networks:
- It replicates the communication for multi-node WBANs by molecular communication.
Tools and Resources
- Nano-Sim:
- The tool NS3 for extension the nanoscale networks.
- It helps for molecular and electromagnetic nanonetworks.
- Visualization:
- Use the tool python (Matplotlib) or MATLAB for envision the diffusion.
- Research Papers:
- Examine the molecular communication design and protocols for deeper insights.
Here, we completed discussed about Molecular communication that has manage the network to enhance the network in complex scenarios effectively using the ns3 tool. A dedicated manual will be shared to handle further queries about this project.

 Click Here to watch our latest output video using NS3 simulator
Click Here to watch our latest output video using NS3 simulator 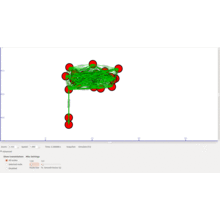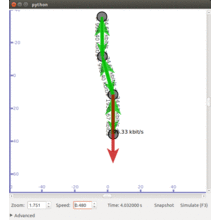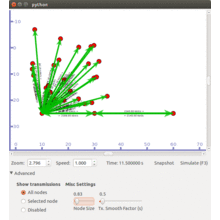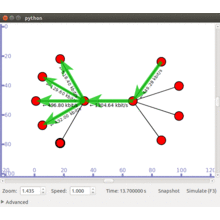 Click Here to watch our latest projects screenshots using NS3 simulator
Click Here to watch our latest projects screenshots using NS3 simulator